Microbiology Lab Notes (media and tests) By ©2018 Shirley S. Chung, Green River Community College
Download these notes.
Thioglycollate Broth:
- Test aerotolerance of bacteria.
- Turns pink in presence of oxygen via Resazurin indicator. Alternatively, methylene blue indicator (colorless in an anaerobic environment and greenish-blue in the presence of oxygen).
- Uniform growth = facultative anaerobic bacteria.
- Bubbles = gas-producing bacteria.
- Bottom growth = anaerobic bacteria.
- Contains sodium thioglycollate, thioglycollic acid, L-cystine, methylene blue, and 0.05% agar. The sodium thioglycollate, thioglycollic acid, and L-cystine reduce the oxygen to water.
http://www.austincc.edu/microbugz/fluid_thioglycollate_medium.php
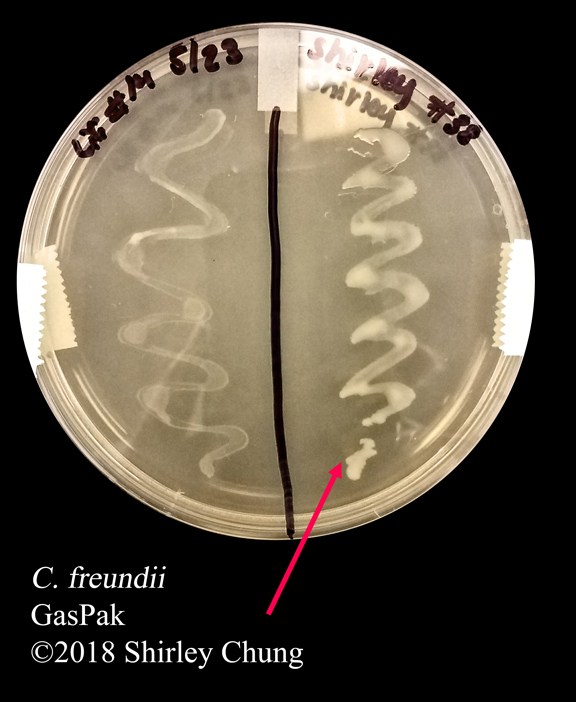
GasPak:
- For incubation of anaerobic cultures in a nonreducing medium.
- Generates water and CO2.
- Methylene blue strip indicator is blue in presence of oxygen, and colorless in absence of oxygen.
Selective, Phenylethyl alcohol agar:
- For isolation of most G+
- Partially inhibitory to G- (may form, but stunted growth, suboptimal).
- Inhibits E. coli, selects for S. aureus.
Selective, Crystal violet agar:
- Selective for most G-
- Inhibitory to most G+
Selective, 7.5% NaCl agar:
- For halophilic
- Inhibitory to most other non-halophilic organisms.
- *Most useful in detection of genus Staphylococcus.
Differential/Selective, MacConkey agar:
- Has bile salts and crystal violet, lactose, neutral red.
- Inhibit G+
- Selective for G-
- Contains Lactose
- Contains pH neutral red which differentiates RED-Lactose-fermenting colonies; translucent-non-fermenting colonies.
- Differentiate between enteric bacteria.
- Coliform bacili: lactose fermenters, make acid, RED color on their surface. coli is a super fermenter and there will be pink zone surrounding growth.
- Dysentery, typhoid, paratyphoid: non-lactose fermenters; TAN appearance or transparent.
Differential/Selective, Mannitol salt agar (MSA):
- High salt concentration, 7.5% NaCl
- Select for staphylococci, inhibit most other non-halophilic bacteria.
- Contains mannitol (carbohydrate) for differential (some staphylococci can ferment).
- Phenol red indicator detect acid from mannitol-fermenting staphylococci.
- Yellow zone around growth is positive for mannitol fermentation; no color change is negative.
Differential/Selective, Eosin-methylene blue agar (Levine):
- Has lactose and dyes eosin and methylene blue.
- Partly inhibitory to G+
- Promote G-
- Differentiate between enteric lactose fermenters and nonfermenters.
- Can identify between E. coli (blue-black w/metallic green sheen due to lots of acid).
- Can identify E. aerogenes (thick mucoid, pink colonies).
- Enteric bacteria that do NOT ferment lactose—colorless colonies, transparent, and appear to take on purple color of medium.
Enriched Media, Blood agar:
- For cultivation of fastidious organisms (e.g. Streptococcus).
- Demonstrate hemolytic properties.
- Gamma: no lysis of RBC. No change in medium.
- Alpha: incomplete lysis of RBC; reduction of Hb to methemoblogin results in greenish halo around growth.
- Beta: lysis of RBC; results in clear zone.
Starch Hydrolysis:
- Extracellular enzyme amylase to hydrolyze starch down to maltose (maltase cat.) then glucose.
- Starch agar.
- Flood with iodine to test: blue-black = presence of starch and NEG for starch hydrolysis; clear zone (exoenzymes present) = POS for starch hydrolysis.
Lipid Hydrolysis:
- Tributyrin agar.
- After inoculation, CLEAR zone POS for lipid hydrolysis (lipase).
Casein Hydrolysis:
- Milk agar to test for protein hydrolysis.
- Clear zone = POS.
Gelatin Hydrolysis:
- Test for liquifaction via gelatinase to hydrolyze protein to amino acids.
- Gel deep tubes get inoculated.
- After incubation, put in fridge for 30min. Cultures that remain liquified produce gelatinase and rapid gelatin hydrolysis (POS).
- If solid, then re-incubate cultures for 5 more days. Put in fridge for 30min. If liquify, then they are POS for SLOW gelatin hydrolysis. If solid, then NEGATIVE.
Carbohydrate Fermentation:
- *Facultative anaerobes are usu. the fermenters.
- Need broth and Durham tube.
- Observe w/in 48 hrs.
- Add phenol red: red turns yellow = POS (no color change of indicator = NEG).
- Gas = POS.
- Beware that neg result does NOT mean no growth.
Hydrogen Sulfide:
- 2 fermenting pathways to produce H2S (g).
- Stab inoculation.
- Black ferrous sulfide = POS.
- Also indicate motility.
Urease:
- Useful to i.d. Proteus vulgaris (produces urease). Other organisms also can produce urease.
- Inoculate urea broth containing indicator phenol red.
- Deep pink = POS urease presence. No deep pink = NEG.
Nitrate Reduction Test:
- Reduction of nitrates by some aerobic/facultative anaerobic organisms occur in absence of oxygen. Use inorganic subtrates NO3 or SO4. Some can further reduce Nitrite to ammonia.
- Solution A=sulfanilic acid. Solution B is alpha-naphthylamine.
- Solution A+B = cherry red = POS for reducing nitrates to nitrites.
- No red gives 2 possibilities: end products were reduced even further down to ammonia; or no reduction took place.
- Add zinc. No color change = no nitrates = POS. Red color change = NEG = yes nitrates are present.
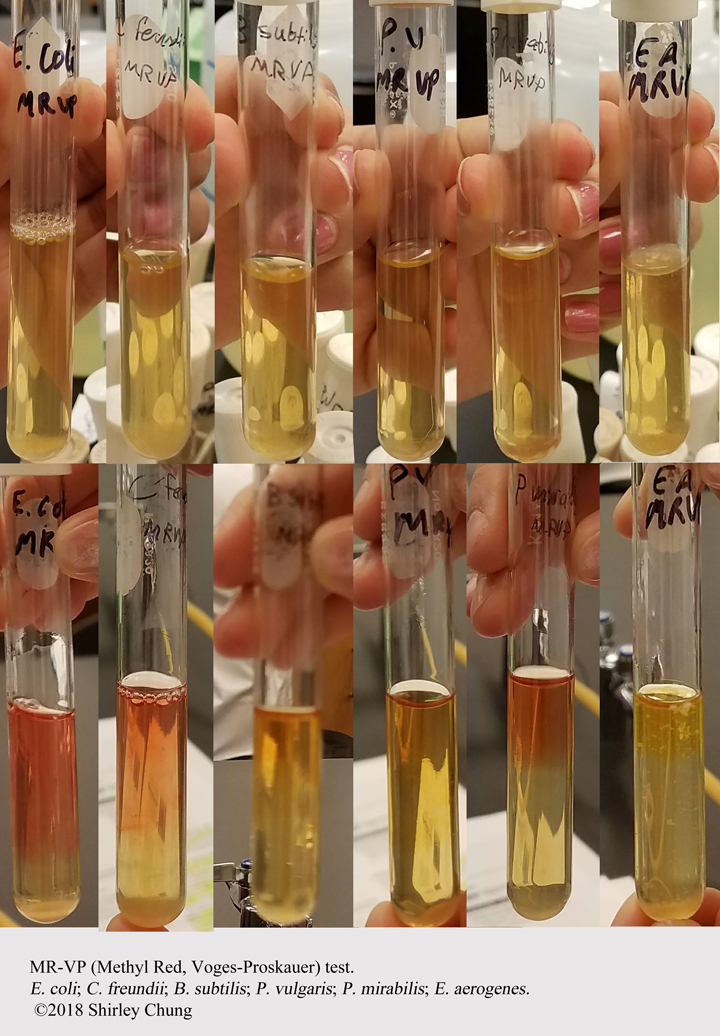
IMViC (Indole, Methyl red, Voges-Proskauer, Citrate utilization):
Barritt’s reagents A+B. Wait 15 min. Rose color = POS (for glucose fermentation). No pink rose color = NEG.
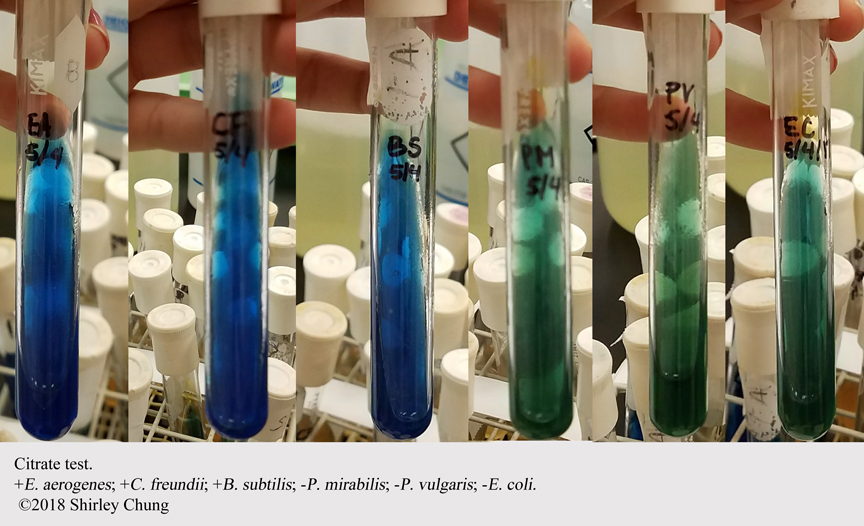
- Citrate Utilization.
Differentiate enteric organisms ability to use citrate as sole source of carbon.
Growth, blue medium = POS for citrate. Green, no growth = NEG for citrate.
Catalase:
- Aerobic respiration, hydrogen peroxide and superoxides are produced. Capable of producing catalase or superoxide dismutase.
- Strict anaerobes don’t produce these enzymes.
- Differentiate catalase-positive Staphylococci and catalase-negative Streptococci and members of Enterobacteria.
- Inoculate on TSA slant/plate.
- Add H2O2 (hydrogen peroxide). Bubbles = POS for catalase. No bubbles = NEG = strict anaerobe.
Oxidase:
- Aerobic bacteria and some facultative exhibit oxidase activity.
- Differentiate between Neisseria and Pseudomonas (both oxidase-positive) and Enterobacteria (oxidase-negative).
- Test reagent alpha-aminodimethylaniline.
- Pink then maroon then dark purple = POS for cytochrome oxidase production.
- No color change or light pink = NEG.
Microbiology Lab Procedure Notes By ©2018 Shirley S. Chung, Green River Community College